
Up: Hydractinia
Previous: Results
Generally, we were fairly pleased with the resulting dynamics of our model - they mimicked those described in the assignment to a suitable degree of accuracy, and provided a few interesting and unexpected results. In particular, the buffering phenomenon was an unexpected (though in retrospect, understandable) action in the given model. Even more interesting, however, was the change in the buffering phenomenon with the activation of genotypic mutation. Although it would be presumptuous to claim that the colonies were knowingly doing anything, genotypic mutation provided a clear competitive advantage to all colonies. Those which were stifled by buffering were permitted the opportunity to penetrate the buffer via a mutation which was friendly to the parent colony, but unfriendly to the buffer. Further, colonies being attacked (particularly the fused mat colonies) could survive complete destruction by local mutations which were neutral to both the host colony and the attacking colony.
The model could be improved in many ways, however. First and foremost, the homogeneity of a colony's structure is problematic. In real world Hydractinia colonies are specialized morphological structures which are omitted in our model. The transportation and harnessing of resources was another significant omission. In simulation after simulation, we consistently saw the fused colonies excluded by their less passive morphological counter-parts. Surely, in the real Hydractinia colonies, the fused mat cells serve a more specialized role in conjunction with the other morphologies - the benefits of mat cells promoting increased reproductive potential were not well characterized by the model. In short, while we think that the model did produce some very interesting dynamics, we didn't feel like we were accurately representing the dynamics of real Hydractinia colonies.
Figure 1:
Example of successful allorecognition between a stolon colony (red, genotype:
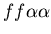 ) and fused mat colony (yellow, genotype:
) and fused mat colony (yellow, genotype:
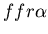 ). Images are at timesteps 20, 70, 100, 150, 300 and 5000. In the final snapshot, we can clearly see that the two colonies are diffusing amongst each other to some degree, as expected with fused colonies.
). Images are at timesteps 20, 70, 100, 150, 300 and 5000. In the final snapshot, we can clearly see that the two colonies are diffusing amongst each other to some degree, as expected with fused colonies.
|
|
Figure 2:
Example of passive rejection by two fused mat colonies (genotypes:
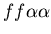 and
and
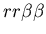 ). Images are from timesteps 40, 90, 140, 190, 250 and 600. Not the lack of dead cells (black) at the intersection of the two colonies - this indicates that there is no fighting between the colonies.
). Images are from timesteps 40, 90, 140, 190, 250 and 600. Not the lack of dead cells (black) at the intersection of the two colonies - this indicates that there is no fighting between the colonies.
|
|
Figure 3:
Example of active rejection by a stolon colony (red, genotype:
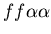 ) and a fused mat colony (yellow, genotype:
) and a fused mat colony (yellow, genotype:
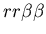 . Images are from timesteps 50, 130, 190, 230, 350 and 600. Note the thin region of dead cells (black) which persists at the intersection of the two colonies; this region indicates active rejection resulting in the death of cells (fused mat cells, in this case).
. Images are from timesteps 50, 130, 190, 230, 350 and 600. Note the thin region of dead cells (black) which persists at the intersection of the two colonies; this region indicates active rejection resulting in the death of cells (fused mat cells, in this case).
|
|
Figure 4:
Example of active rejection between a stolon colony (red, genotype:
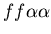 ) and an unfused mat colony (blue, genotype:
) and an unfused mat colony (blue, genotype:
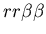 ). The unfused mat colony grows at a higher rate than the stolon colony as a result of the presence of other mat cells increasing the probability of reproduction. Images are from timesteps 50, 90, 140, 200, 500 and 920.
). The unfused mat colony grows at a higher rate than the stolon colony as a result of the presence of other mat cells increasing the probability of reproduction. Images are from timesteps 50, 90, 140, 200, 500 and 920.
|
|
Figure 5:
Example of the buffering phenomenon, in which the fighting region between two hostile colonies is colonized by a third colony which is neutral to both aggressors, resulting in a stable, non-hostile arrangement of cells. The two hostile colonies are unfused mat colonies (light and dark blue, genotypes:
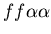 and
and
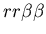 , while the buffering colony is a stolon (red, genotype:
, while the buffering colony is a stolon (red, genotype:
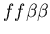 ). Images are from timesteps 70, 150, 200, 350, 700 and 1500.
). Images are from timesteps 70, 150, 200, 350, 700 and 1500.
|
|
Figure 6:
Example of a single colony (unfused mat, blue, genotype:
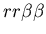 ) dominating others (stolon and fused mat, red and yellow, genotypes (
) dominating others (stolon and fused mat, red and yellow, genotypes (
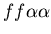 and
and
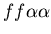 ) via competitive exclusion. The unfused mat colony, being significantly larger, is able to slowly whittle down the resistant stolon colony. Images are from timesteps 70, 120, 180, 300, 600 and 5000.
) via competitive exclusion. The unfused mat colony, being significantly larger, is able to slowly whittle down the resistant stolon colony. Images are from timesteps 70, 120, 180, 300, 600 and 5000.
|
|
Figure 7:
Rock-Paper-Scissors experiment (as suggested in the assignment), in which two colonies fuse (unfused mat and stolon, blue and red, genotypes:
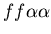 and
and
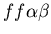 ), while one rejects (stolon, red, genotype:
), while one rejects (stolon, red, genotype:
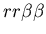 ).
).
|
|
Figure 8:
Activating genotypic mutation for the system used in Figure 6 (example 1). Images are timesteps 50, 140, 250, 380, 450 and 800.
|
|
Figure 9:
Activating genotypic mutation for the system used in Figure 6 (example 2). Images are timesteps 110, 320, 410, 500, 640 and 1000.
|
|


Up: Hydractinia
Previous: Results
THIS DOCUMENT HAS NOT BEEN REVIEWED -- PLEASE DO NOT CITE